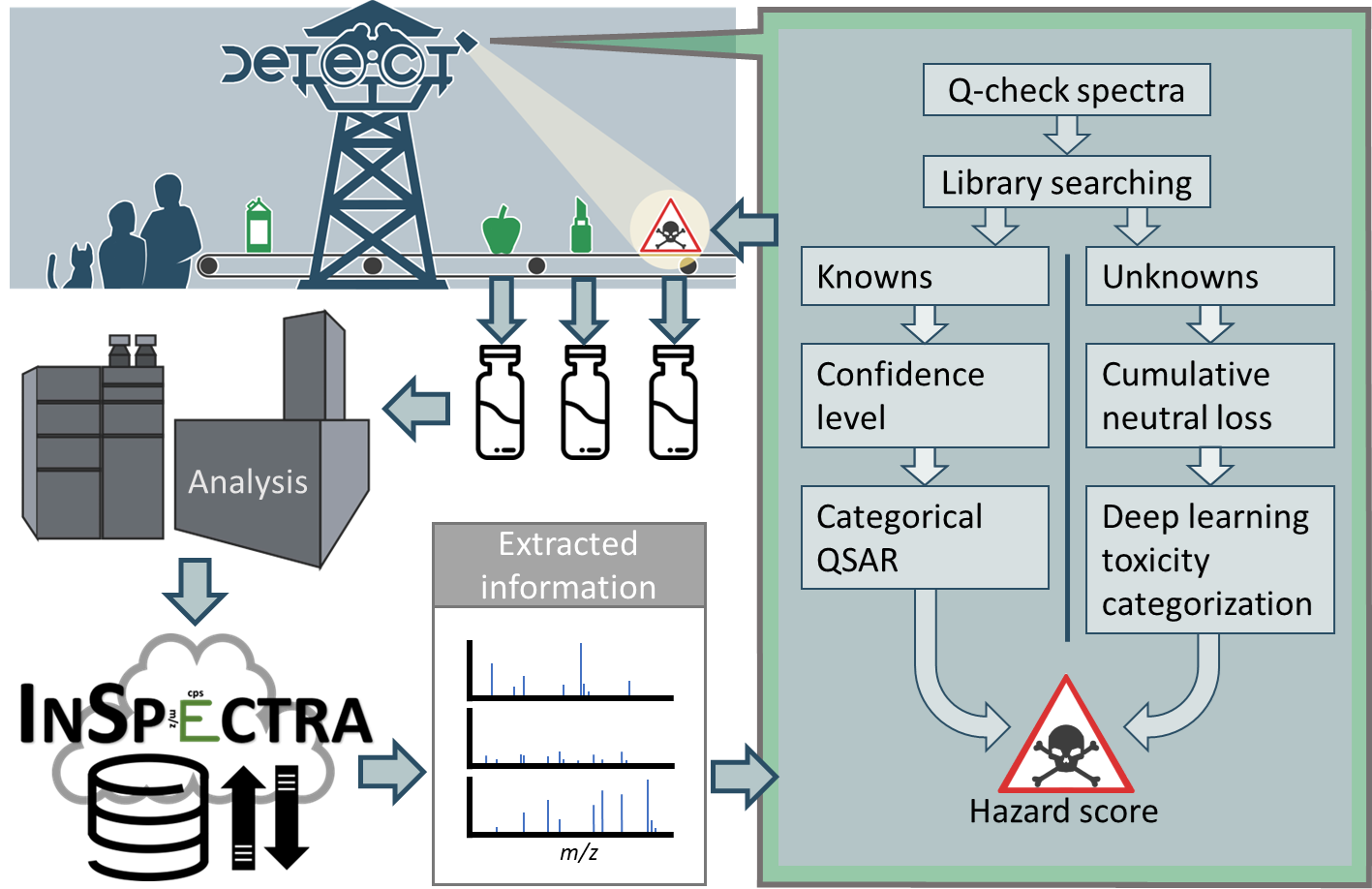
Ongoing Projects
DETECT
from HRMS Detection to Environmental Contamination Trigger
Dr. Saer Samanipour (PI), Dr. Lapo Renai (postdoc), Viktoriia Turkina (PhD candidate), Tobias Hulleman (PhD candidate), Xander Becking (PhD candidate)
DETECT (Data-Driven Environmental Chemistry Toolset) is a research project focused on developing predictive models using high-resolution mass spectrometry (HRMS) data to better understand and map the chemical space of the exposome. Instead of aiming for chemical identification, DETECT focuses on extracting patterns and building models that relate spectral features to underlying physicochemical properties and environmental behavior. The project leverages non-targeted analysis (NTA) datasets and mass spectral fragmentation data to train machine learning algorithms that predict descriptors such as chromatographic retention, polarity, volatility, and molecular class. These models help researchers understand the structure–response relationships in environmental samples and enable a more robust exploration of chemical space, even when the compounds are unknown or absent from spectral libraries. DETECT aims to move beyond traditional database matching by applying data-driven approaches that capture the diversity of real-world environmental mixtures. The insights gained from these models support risk prioritization, chemical grouping, and a better mechanistic understanding of how chemicals behave in the environment—without requiring full structural elucidation. The tools and models developed through DETECT contribute to open science initiatives and are integrated into broader exposome research frameworks, ultimately enabling more informed decision-making in environmental monitoring and policy.
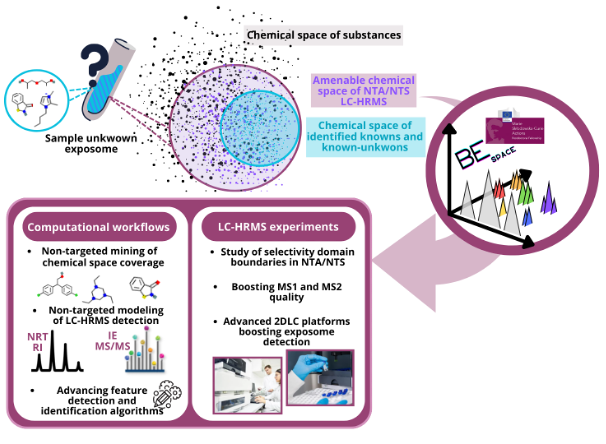
BESPACE
BEspace: Boosting the exposome space coverage in the aquatic environment by multidimensional integrated analytical platform
Dr. Saer Samanipour (PI) and Dr. Lapo Renai (postdoc)
BEspace is an MSCA funded project focusing on enhancing the detection capabilities of non-targeted analysis and screening of the exposome chemical space in aquatic environments. The project aims to develop and apply advanced computational (i.e., machine learning-based tools) and analytical (i.e., two-dimensional liquid chromatography) strategies to improve the accuracy and quality of detected known and unknown chemicals, ultimately contributing to more realistic and conservative non-targeted environmental monitoring.
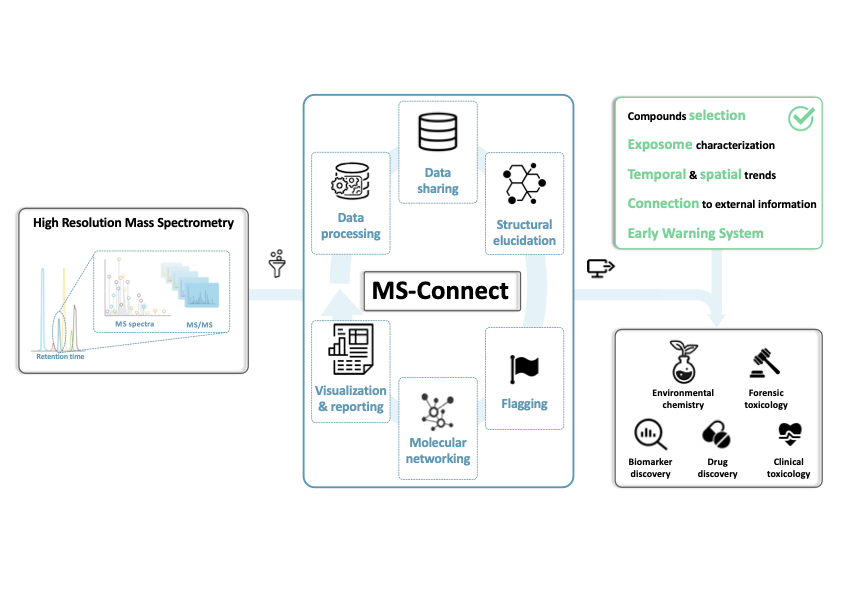
MS-Connect
an open access platform for complete mining of high-resolution mass spectrometry data
MS-Connect is a research project focused on the development of digital infrastructure and computational tools to map and identify the measured exposome chemical space from non-targeted high-resolution mass spectrometry (HRMS) data. As environmental samples contain thousands of unknown features, traditional chemical identification approaches fall short in capturing the full complexity and diversity of real-world exposures. The primary goal of MS-Connect is to bridge analytical chemistry and data science by building scalable tools that connect mass spectral features to known and unknown chemical structures. The project integrates spectral similarity networks, cheminformatics-based annotation workflows, and machine learning models to systematically organize detected signals and prioritize those most relevant to environmental and human health. By creating interactive and interoperable data platforms, MS-Connect enables the visualization and exploration of the exposome chemical space, facilitating both structural elucidation and hypothesis generation. The tools developed within this project are designed to support researchers, regulators, and data curators in linking chemical fingerprints to potential sources, pathways, and effects. MS-Connect plays a key role in ongoing exposome research efforts by providing community-accessible, open-source solutions that advance the field of environmental mass spectrometry and chemical knowledge discovery.
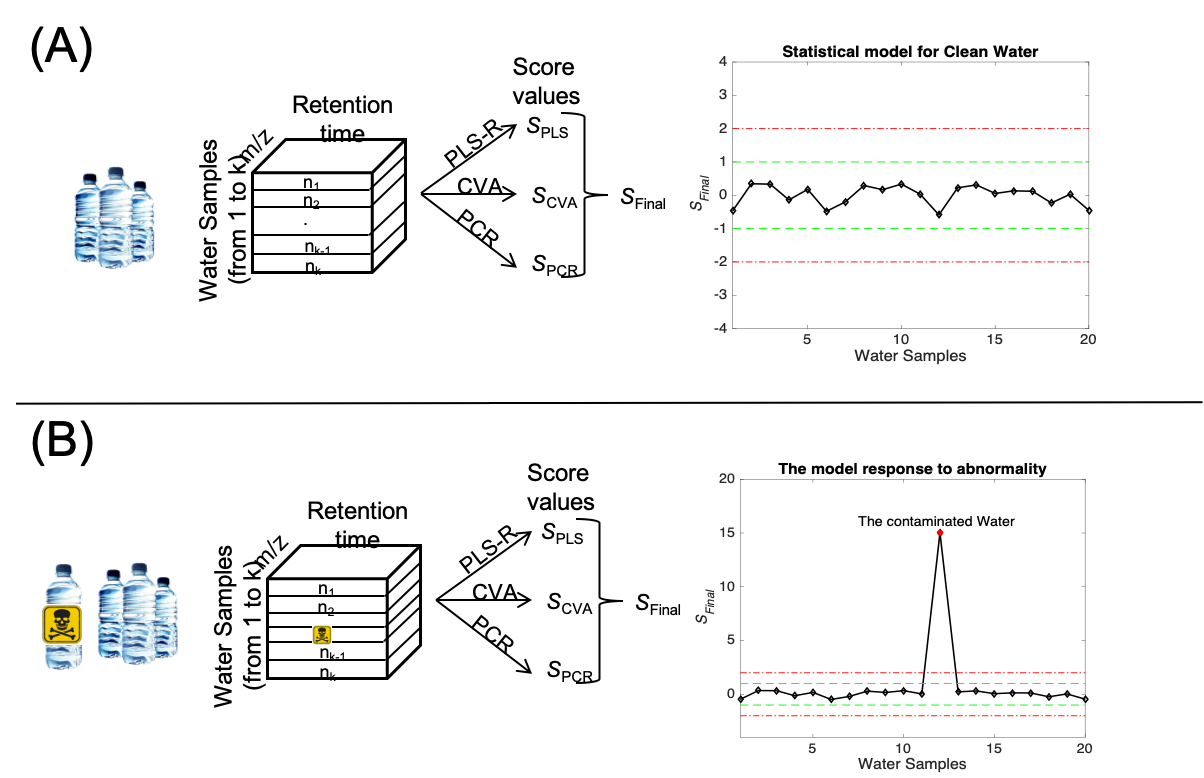
SCOPE
Smart analysis of Complex Organic mixtures in Pre-biotic Environments
SCOPE is a research project dedicated to developing data-driven methods for detecting chemical anomalies in complex environmental and biological samples using non-targeted high-resolution mass spectrometry (HRMS). In the context of exposomics, these anomalies may represent rare exposures, emerging contaminants, or unexpected transformation products that fall outside typical chemical patterns. The project focuses on statistical and machine learning techniques for identifying features that deviate from expected chemical behavior across large and diverse sample sets. SCOPE aims to establish robust baseline models of chemical occurrence and variability, which serve as references for detecting unusual signals—both structurally and statistically. Key goals include the detection of outliers in spectral feature distributions, the identification of temporal or spatial chemical shifts, and the classification of sample types based on anomalous chemical fingerprints. By combining unsupervised learning, feature prioritization, and chemical metadata analysis, SCOPE contributes to the early discovery of emerging chemical threats and supports exploratory environmental research. The outputs of SCOPE include open-source analytical workflows, anomaly detection frameworks tailored for mass spectrometry data, and data visualization tools that enable researchers and regulators to focus on the most atypical and potentially high-impact signals in the exposome.